Table 2. Summary statistics of the utilized genetic diversity parameters in the study.
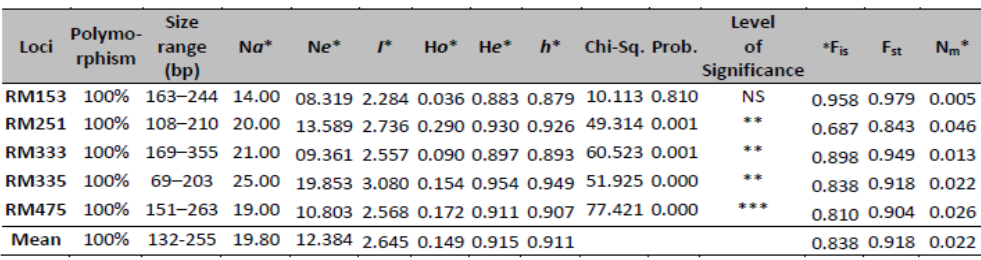
*Na = Number of observed alleles, *Ne = Number of effective alleles =1/(∑Xi2), Where Xi is the frequency of the ith allele [43], *I = Shannon's Information index by Lewontin [44], *Unbiased Expected Heterozygosity (He) was computed using Levene [45], *h = Nei's (1973) gene diversity index [46], NS=not significant, * P<0.05, ** P<0.01, *** P<0.001, *Fis = Wright's fixation index (Fis) as a measure of heterozygote deficiency or excess (He - Ho) / He) [47], and *Nm = Gene flow estimated from Fst = 0.25(1 - Fst)/Fst